Sparse graphs from line graphons
linegraphons.Rmd
library(graphonmix)
library(ggplot2)
library(igraph)
#>
#> Attaching package: 'igraph'
#> The following objects are masked from 'package:stats':
#>
#> decompose, spectrum
#> The following object is masked from 'package:base':
#>
#> union
requireNamespace("gridExtra", quietly = TRUE)Line graphs
Suppose is a graph. Then the line graph of , commonly denoted by maps edges of to vertices of and two vertices in are connected if the original edges in share a common vertex. Here’s an example.
g1 <- make_star(4,mode = "undirected")
g1 <- add_vertices(g1, 1)
g1 <- add_edges(g1,c(4,5))
lg1 <- make_line_graph(g1)
oldpar <- par(mfrow = c(1,2))
plot(g1, vertex.label = NA, edge.label = c(1,2,3,4), main = "Graph G")
plot(lg1, vertex.label = c(1,2,3,4), vertex.size = 20, main = "Line graph L(G)")
par(oldpar)Line graphs of star graphs
The key is that line graphs of stars are complete graphs. In the example below each edge of graph is a vertex in . Two vertices in are connected if the corresponding edges in share a vertex.
st <- make_star(10, mode = "undirected")
lgr_st <- make_line_graph(st)
oldpar <- par(mfrow = c(1,2))
plot(st, vertex.label = NA, main = "Star graph G")
plot(lgr_st, vertex.label = NA, main = "Line graph L(G) of a star")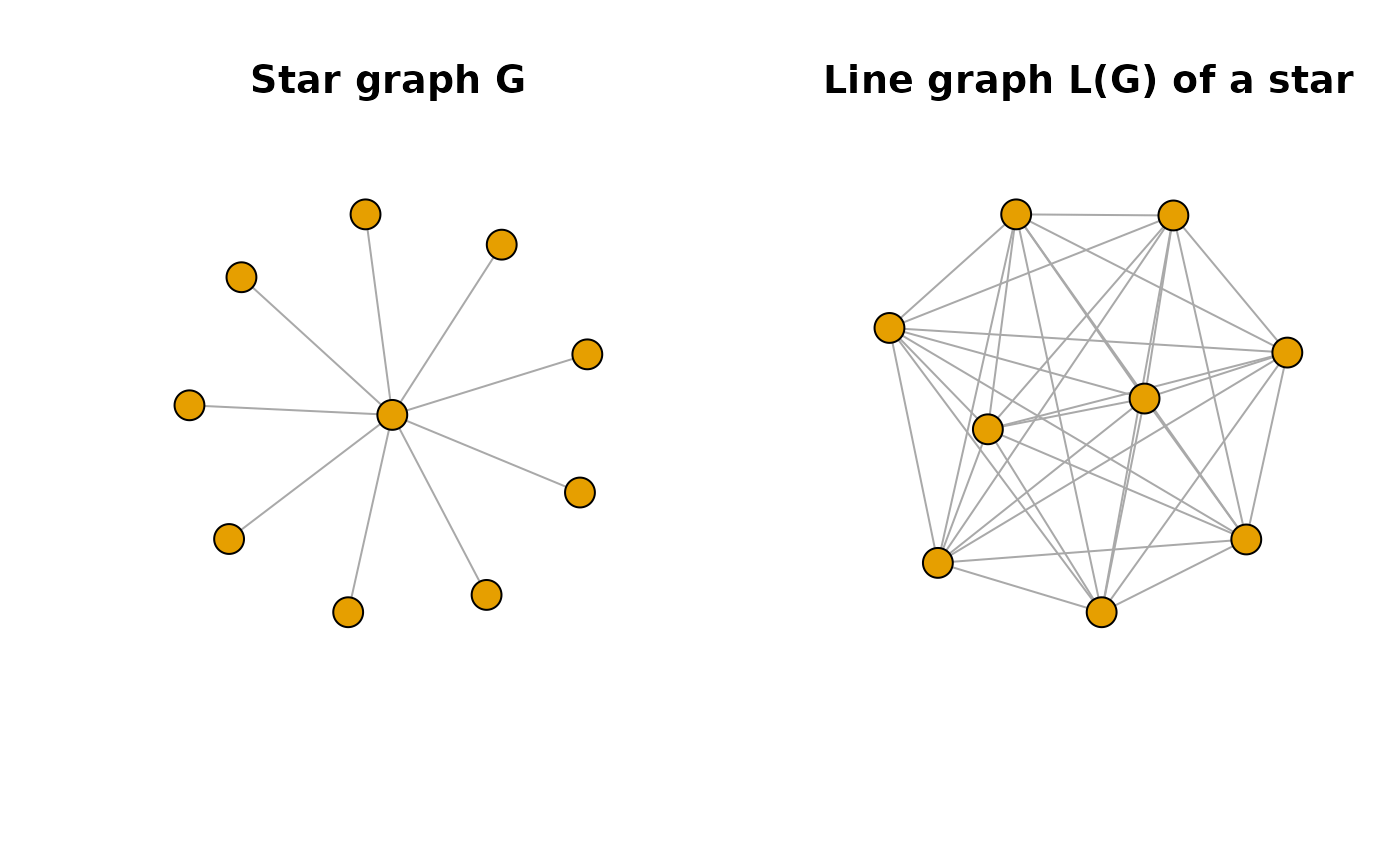
par(oldpar)Sparse graphs
A sequence of graphs is sparse if the edges grow subquadratically with the nodes. Star graphs are the ultimate sparse graphs, because a star of nodes has edges. A graph sequence is dense if the edges grow quadratically with the nodes. Erdos-Renyi graphs with a fixed edge probability are dense. This can be seen from the edge density. Let’s generate a sequence of star graphs and a sequence of Erdos-Renyi graphs and see this in action.
stardensity <- gnpdensity <- rep(0, 20)
for(i in 1:20){
n <- i*100
gr1 <- sample_gnp(n, p = 0.1)
gnpdensity[i] <- edge_density(gr1)
gr2 <- make_star(n, mode = "undirected")
stardensity[i] <- edge_density(gr2)
}
gnpdensity
#> [1] 0.09696970 0.10120603 0.09895206 0.09996241 0.09983166 0.10017807
#> [7] 0.09997956 0.09944931 0.10038561 0.09987988 0.10030275 0.09965388
#> [13] 0.09979392 0.10010926 0.09974917 0.10025094 0.09994807 0.09999568
#> [19] 0.10022560 0.10025063
stardensity
#> [1] 0.020000000 0.010000000 0.006666667 0.005000000 0.004000000 0.003333333
#> [7] 0.002857143 0.002500000 0.002222222 0.002000000 0.001818182 0.001666667
#> [13] 0.001538462 0.001428571 0.001333333 0.001250000 0.001176471 0.001111111
#> [19] 0.001052632 0.001000000We see that the density of stars goes to zero whereas the density of GNP graphs hover around 0.1, because we have used the edge probability as 0.1.
Graphons
Graphons are graph limits. Suppose we have a graph . The empirical graphon of is when we scale the adjacency matrix to the unit square and color the small squares that correspond to ones in the adjacency matrix in black. Here we plot the empirical graphon of a star graph with 10 nodes.
gr <- make_star(10, mode = "undirected")
emp <- empirical_graphon(gr)
plot_graphon(emp) + coord_fixed()
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the graphonmix package.
#> Please report the issue to the authors.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.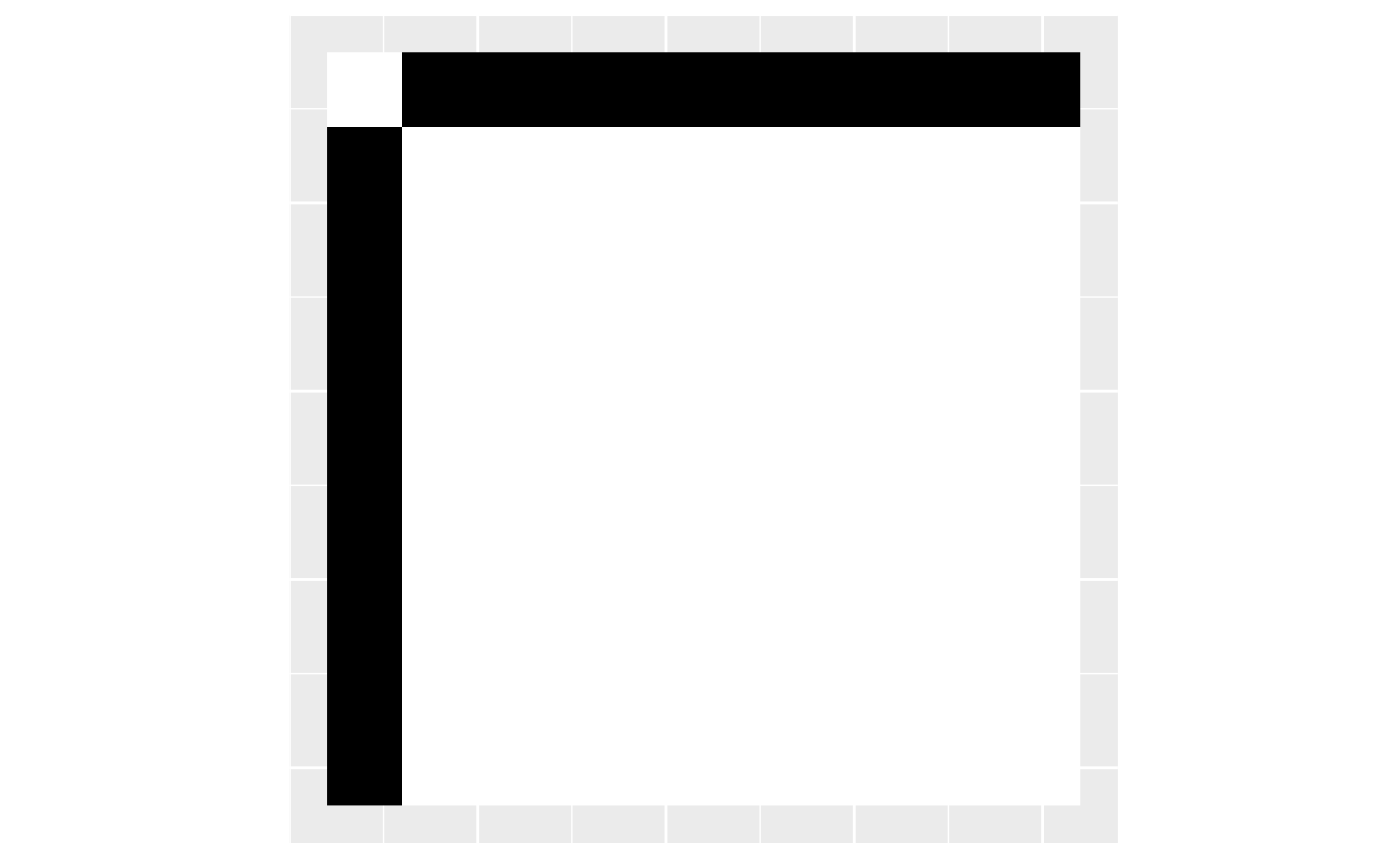
Graphons of sparse graphs
The problem is graphons of sparse graphs are zero. Graphons are generally denoted by the letter . And if the graphs are sparse then . That is, the empirical graphons converge to zero. Here, we’re a bit handwavy about convergence (actually, the convergence is with respect to the cut metric, but let’s let’s not discuss that).
gr <- make_star(20, mode = "undirected")
emp <- empirical_graphon(gr)
g1 <- plot_graphon(emp) + coord_fixed() + ggtitle('n = 10')
gr <- make_star(100, mode = "undirected")
emp <- empirical_graphon(gr)
g2 <- plot_graphon(emp) + coord_fixed() + ggtitle('n = 100')
gr <- make_star(200, mode = "undirected")
emp <- empirical_graphon(gr)
g3 <- plot_graphon(emp) + coord_fixed() + ggtitle('n = 200')
gridExtra::grid.arrange(g1, g2, g3, nrow = 1)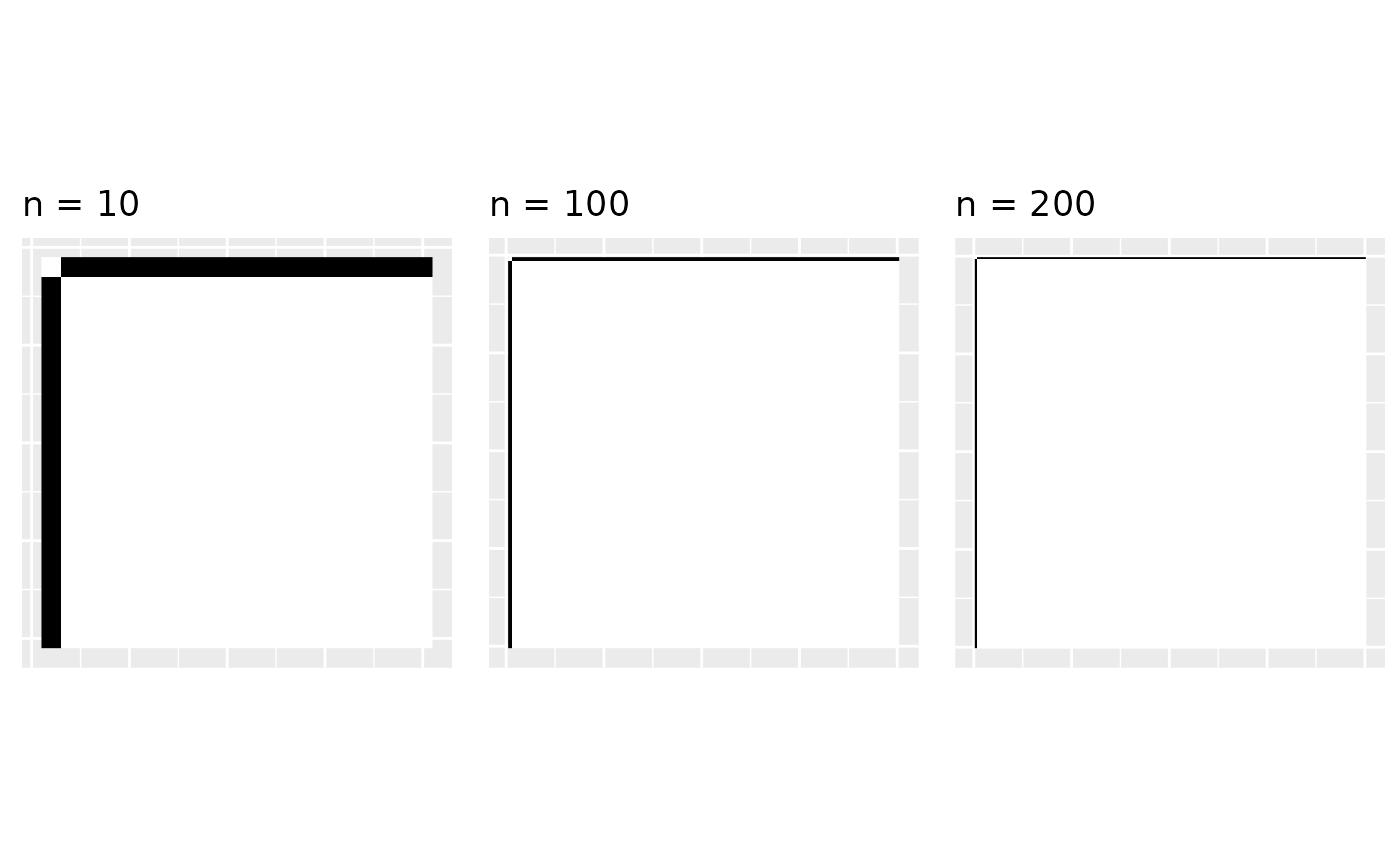
As , there are less black parts in the empirical graphon and it converges to zero. When the graphon is zero, we cannot sample from it; it is not useful anymore. So what do we do?
Graphons of line graphs
For a subset of sparse graphs, such as the star graphs, their line graphs are dense. We discuss this in detail in (Kandanaarachchi and Ong 2024). The main example is star graphs.
gr1 <- make_star(10, mode = "undirected")
gr <- gr1 %du% gr1
lgr <- make_line_graph(gr)
emp <- empirical_graphon(lgr)
g1 <- plot_graphon(emp) + coord_fixed() + ggtitle('n = 20')
gr1 <- make_star(50, mode = "undirected")
gr <- gr1 %du% gr1
lgr <- make_line_graph(gr)
emp <- empirical_graphon(lgr)
g2 <- plot_graphon(emp) + coord_fixed() + ggtitle('n = 100')
gr1 <- make_star(100, mode = "undirected")
gr <- gr1 %du% gr1
lgr <- make_line_graph(gr)
emp <- empirical_graphon(lgr)
g3 <- plot_graphon(emp) + coord_fixed() + ggtitle('n = 200')
gridExtra::grid.arrange(g1, g2, g3, nrow = 1)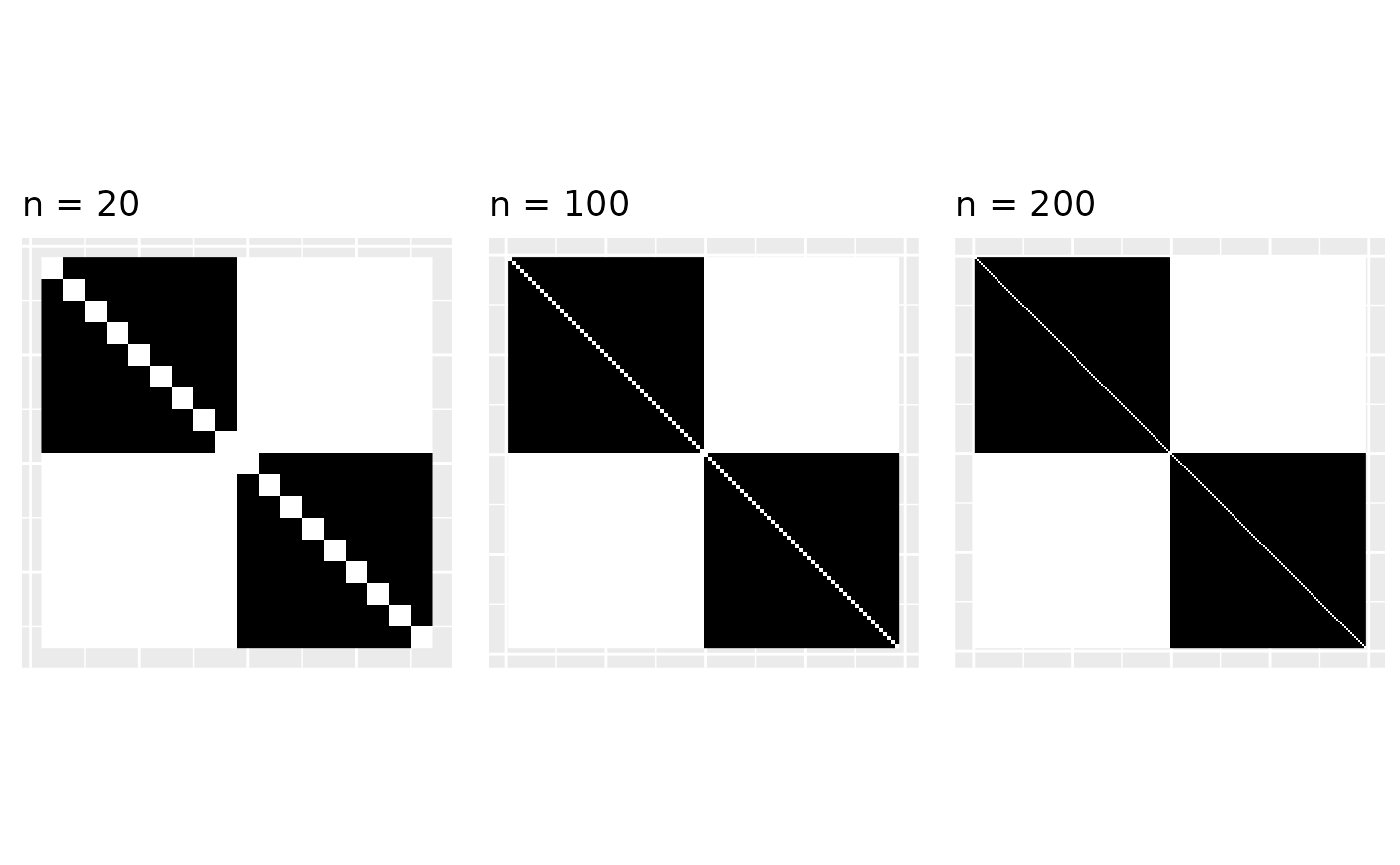
For the above example of two stars, the line graphs are dense and converge to a non-zero graphon.
Janson’s theorem
Janson (Janson 2016) proved that all line graph limits are disjoint clique graphs. This is a very important theorem for us because it tells us what the structure of line graph limits are. Janson explains that line graph limits can be written as a sequence of numbers such that and . The gives the width of each black box in the graphon.
Generating stars as inverse line graphs
From a line graph limit we can generate a disjoint union of clique graphs like this.
wts <- c(0.5, 0.3, 0.2)
U <- line_graphon(wts)
gr <- sample_graphon(U, n = 100)
plot(gr,
vertex.size = 1,
edge.color = "lightgray", # Light colored edges
vertex.label = NA,
vertex.color = "lightblue"
)
The inverse line graphs of disjoint clique graphs are star graphs. And star graphs are sparse. We can generate star graphs using the weights or the partition .
grsp <- generate_star_union(wts, n = 100)
plot(grsp,
edge.curved = 0.3,
vertex.size = 1,
edge.color = "lightgray", # Light colored edges
vertex.label = NA,
vertex.color = "lightblue"
)
This is how the sparse part of the -mixture graph is generated. The vignette at Articles/Getting Started explains the mixture procedure.